
Dimensionality reduction is one of techniques of transformations in unsupervised learning. If you have a lot of features in dataset, for example breast cancer dataset has 30 features, it could be essential to extract or compute information of some kind from dataset. The primary goal of dimensionality reduction techniques is to abstract data characteristis and extract information of some kind.
In my understanding we can use the dimensionality reduction techniques for some purposes as below.
- To extract informative features that could be used in the later learning
- To achieve more faster computation with less dimensions for a model
- To draw a scatter plot in 2D dimensional space with transformed dataset
In this guide, I covered 3 dimensionality reduction techniques 1) PCA (Principal Component Analysis), 2) MDS, and 3) t-SNE for the Scikit-learn breast cancer dataset. Here’s the result of the model of the original dataset. The test accuracy is 0.944 with Logistic Regression in the default setting.
Logreg Train Accuracy: 0.948
Logreg Test Accuracy: 0.944
Logreg Train F1 Score: 0.959
Logreg Test F1 Score: 0.955PCA (Principal Component Analysis)
According to Wikipedia, Principal component analysis (PCA) is the process of computing the principal components and using them to perform a change of basis on the data, sometimes using only the first few principal components and ignoring the rest. We might say that PCA leaves the most informative initial dimensions that are called principal components. Those information can capture most of the variation in the original dataset.
I don’t have enough experiences to understand math behind the PCA. This operation is a part of matrix factorization that is similar to SVD (Singular Value Decomposition) in linear algebra area. Let’s standarize (z-score normalization) the dataset and apply PCA with 2 x n_components option. I visuzlied the transformed data in the 2 dimension scatter plot and confirmed how accuracy changed from the original dataset model.
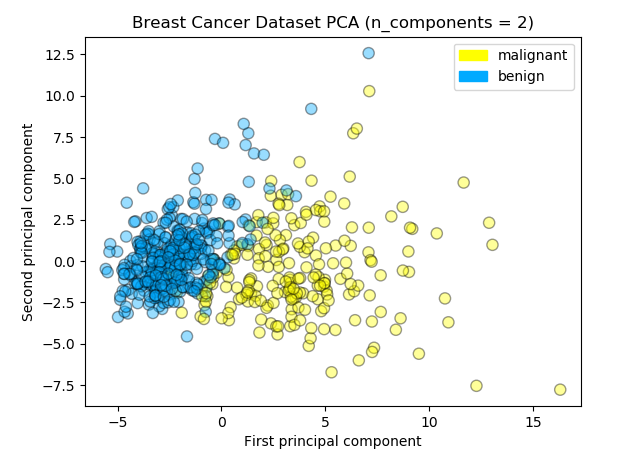
Logreg Train Accuracy: 0.962
Logreg Test Accuracy: 0.951
Logreg Train F1 Score: 0.970
Logreg Test F1 Score: 0.960We’ve got the accuracy for test data 0.951 and saw a little improvement. The projected data points in the scatter plot seem to be clustered into 2 groups and be separated by the boundary straight line. If you install the mglearn library you can draw the scatter plot and the separator easily like illustrated below.

We can create a heat map that visualizes the first two principal components of the breast cancer dataset to get an idea of what feature groupings each component is associated with. PCA.components_ attribute gives us the arrays representing the principal component axes with the amount of variance of each feature. Here’s the visualization of PCA.components_ with matplotlib.

PCA gives a good initial tool for exploring a dataset, but may not be able to find more subtle groupings that produce better visualizations for more complex datasets. There is a family of unsupervised algorithms called Manifold Learning Algorithms that are very good at finding low dimensional structure in high dimensional data and are very useful for visualizations.
MDS (Multi-dimensional Scaling)
PCA is a great tool to start feature engineering on your dataset because it’s fast and interpritable however it’s not much powerful when we want to find non-linear structure in dataset. Here’s the place non-linear dimensionality reduction approaches take place. Manifold learning is one of the major approaches to non-linear dimensionality reduction. Algorithms for this task are based on the idea that the dimensionality of many data sets is only artificially high.
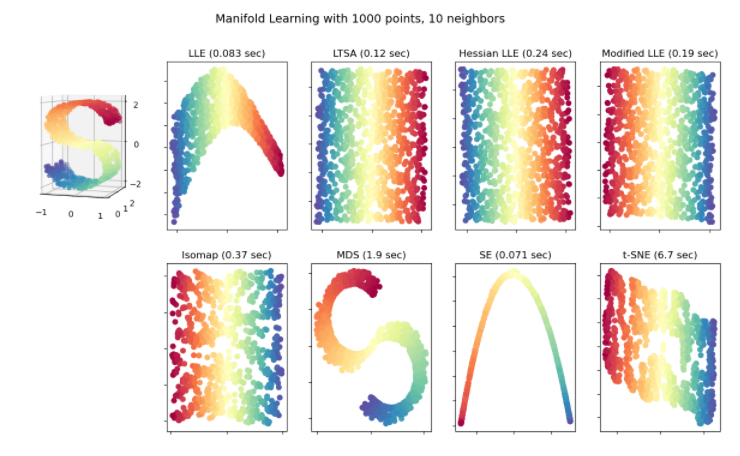
As you see in the below illustration, there are a couple of methods we can apply in Manifold learning such as: Multi-dimensional Scaling (MDS), Locally Linear Embedding (LLE), and Isometric Mapping (IsoMap). I tested 2 of them MDS and t-SNE with the breast cancer dataset in the following section. You can find this illustration from Scikit-learn Manifold learning page. Let’s move to the first approach Multi-dimensional Scaling (MDS).
When you think of a manifold, I’d suggest imagining a sheet of paper: this is a two-dimensional object that lives in our familiar three-dimensional world, and can be bent or rolled in that two dimensions. In the parlance of manifold learning, we can think of this sheet as a two-dimensional manifold embedded in three-dimensional space.
Manifold learning
Multi-dimensional Scaling is one widely used manifold learning methods. There are some variants of MDS, but they all have the same general goal; to seek a low-dimensional representation of the data in which the distances respect well the distances in the original high-dimensional space. In most cases it is useful to find a two-dimensional page – in a way that preserves information about how the points in the original data space are close to each other.
Scikit-learn manifold.MDS implements both types of MDS algorithms: metric and non metric. In default the metric option is set True so metric algorithm was used for this experiment.
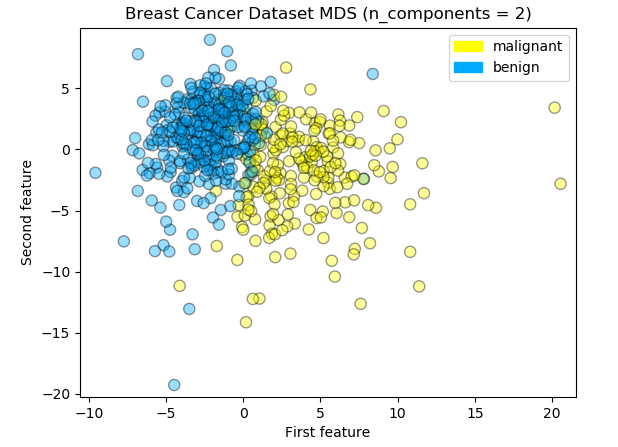
Logreg Train Accuracy: 0.960
Logreg Test Accuracy: 0.930
Logreg Train F1 Score: 0.968
Logreg Test F1 Score: 0.945The accuracy for the test data lowered to 0.93 and there are some distant outliers from the decision boundary. There are some triangles in the right area while some blue circles in the left area. I think the linear dimensionality reduction technique such as PCA was just fine for the breast cancer dataset from the result.

t-SNE
An especially powerful manifold learning algorithm for visualizing your data is called t-SNE. t-SNE finds a two-dimensional representation of your data, such that the distances between points in the 2 dimensional scatter plot match as closely as possible the distances between the same points in the original high dimensional dataset. In particular, t-SNE gives much more weight to preserving information about distances between points that are neighbors. There is detailed information about t-SNE in Scikit-learn documentation.
t-SNE (TSNE) converts affinities of data points to probabilities. The affinities in the original space are represented by Gaussian joint probabilities and the affinities in the embedded space are represented by Student’s t-distributions. This allows t-SNE to be particularly sensitive to local structure and has a few other advantages over existing techniques.
Manifold learning
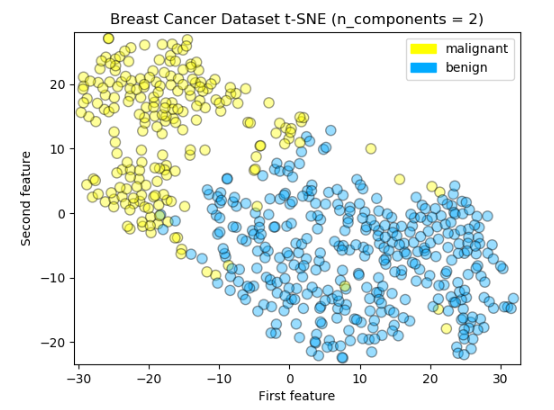
Logreg Train Accuracy: 0.960
Logreg Test Accuracy: 0.930
Logreg Train F1 Score: 0.968
Logreg Test F1 Score: 0.944The interesting thing here is that both MDS and t-SNE did not do better job of finding structure in this rather small and simple breast cancer dataset. It depends on datasets characteristics, dense or sparse, how high dimensional then it can be determined which dimensionality reduction technique can be applied correctly and we will know if it would work bettter or not. For example t-SNE tends to work better on datasets that have more well-defined local structure; in other words, more clearly defined patterns of neighbors.

In our test case, PCA worked well and fastest to do dimensionality reduction. Also it performs well when we trained datasets into our model from accuracy standpoint. We need to observe datasets carefully and test some dimansionality reduction techniques to find the best solution for the transformed data representing infromative features for model training (if your main purpose was to use low dimensional dataset for later learning).
3 ways to do dimensional reduction techniques in Scikit-learn